随机森林算法(Random Forest)R语言实现
随机森林
- 1. 使用Boston数据集进行随机森林模型构建
- 2. 数据集划分
- 3.构建自变量与因变量之间的公式
- 4. 模型训练
- 5. 寻找合适的ntree
- 6. 查看变量重要性并绘图展示
- 7. 偏依赖图:Partial Dependence Plot(PDP图)
- 8. 训练集预测结果
1. 使用Boston数据集进行随机森林模型构建
library(rio)
library(ggplot2)
library(magrittr)
library(randomForest)
library(tidyverse)
library(skimr)
library(DataExplorer)
library(caret)
library(varSelRF)
library(pdp)
library(iml)
data("boston")as.data.frame(boston)
skim(boston)#数据鸟瞰
plot_missing(boston)#数据缺失
#na.roughfix() #填补缺失
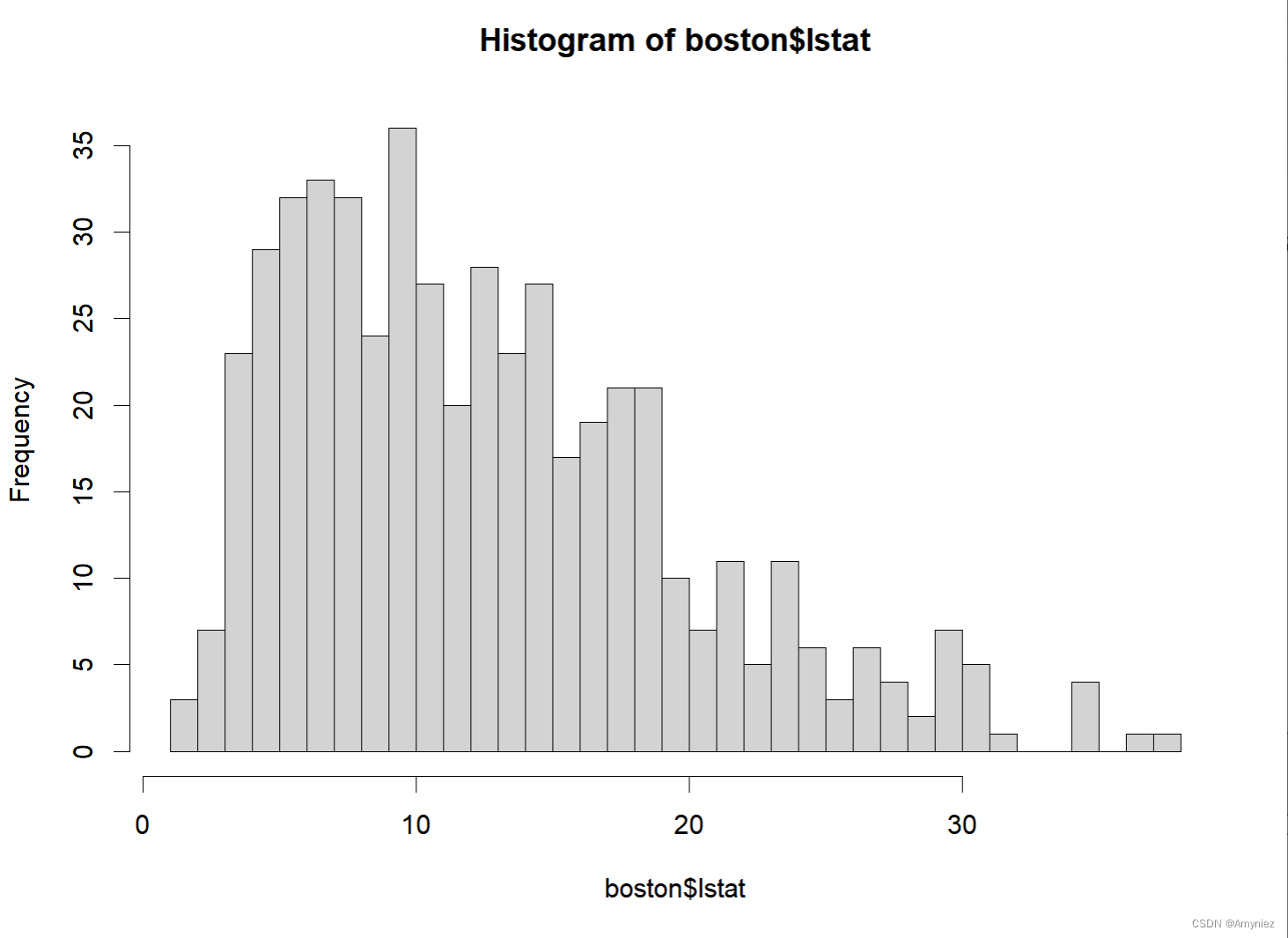
hist(boston$lstat,breaks = 50)
数据展示:
2. 数据集划分
######################################
# 1.数据集划分
set.seed(123)
trains <- createDataPartition(y = boston$lstat,p=0.70,list = F)
traindata <- boston[trains,]
testdata <- boston[-trains,]
3.构建自变量与因变量之间的公式
#因变量自变量构建公式
colnames(boston)
form_reg <- as.formula(paste0("lstat ~",paste(colnames(traindata)[1:15],collapse = "+")))
form_reg

构建的公式:

4. 模型训练
#### 2.1模型mtry的最优选取,mry=12 % Var explained最佳
#默认情况下数据集变量个数的二次方根(分类模型)或1/3(预测模型)
set.seed(123)
n <- ncol(boston)-5
errRate <- c(1) #设置模型误判率向量初始值
for (i in 1:n) {rf_train <- randomForest(form_reg, data = traindata,ntree = 1000,#决策树的棵树p =0.8,mtry = i,#每个节点可供选择的变量数目importance = T #输出变量的重要性)errRate[i] <- mean(rf_train$mse)print(rf_train)
}
m= which.min(errRate)
print(m)
结果:
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 1
Mean of squared residuals: 13.35016% Var explained: 72.5
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 2
Mean of squared residuals: 11.0119% Var explained: 77.31
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 3
Mean of squared residuals: 10.51724% Var explained: 78.33
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 4
Mean of squared residuals: 10.41254% Var explained: 78.55
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 5
Mean of squared residuals: 10.335% Var explained: 78.71
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 6
Mean of squared residuals: 10.22917% Var explained: 78.93
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 7
Mean of squared residuals: 10.25744% Var explained: 78.87
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 8
Mean of squared residuals: 10.11666% Var explained: 79.16
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 9
Mean of squared residuals: 10.09725% Var explained: 79.2
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 10
Mean of squared residuals: 10.09231% Var explained: 79.21
Call:
randomForest(formula = form_reg, data = traindata, ntree = 1000, p = 0.8, mtry = i, importance = T)
Type of random forest: regression
Number of trees: 1000
No. of variables tried at each split: 11
Mean of squared residuals: 10.12222% Var explained: 79.15

结果显示mtry为11误差最小,精度最高
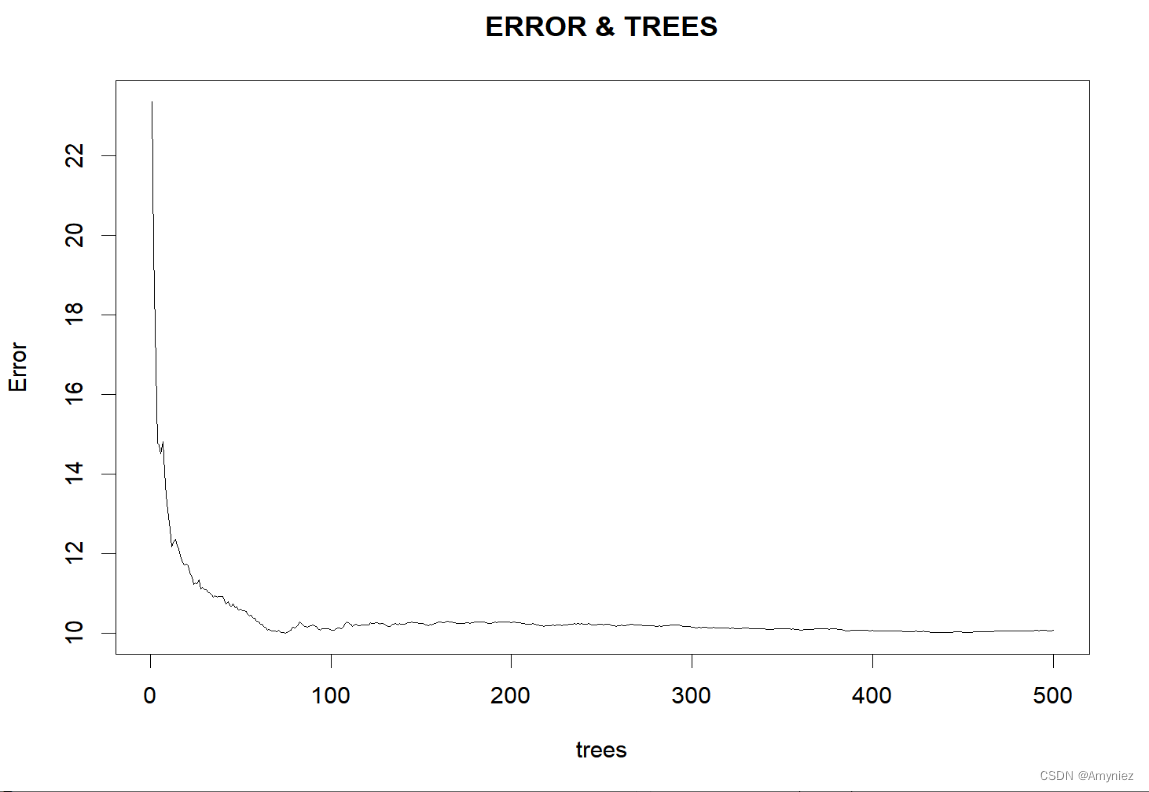
5. 寻找合适的ntree
#### 寻找合适的ntree
set.seed(123)
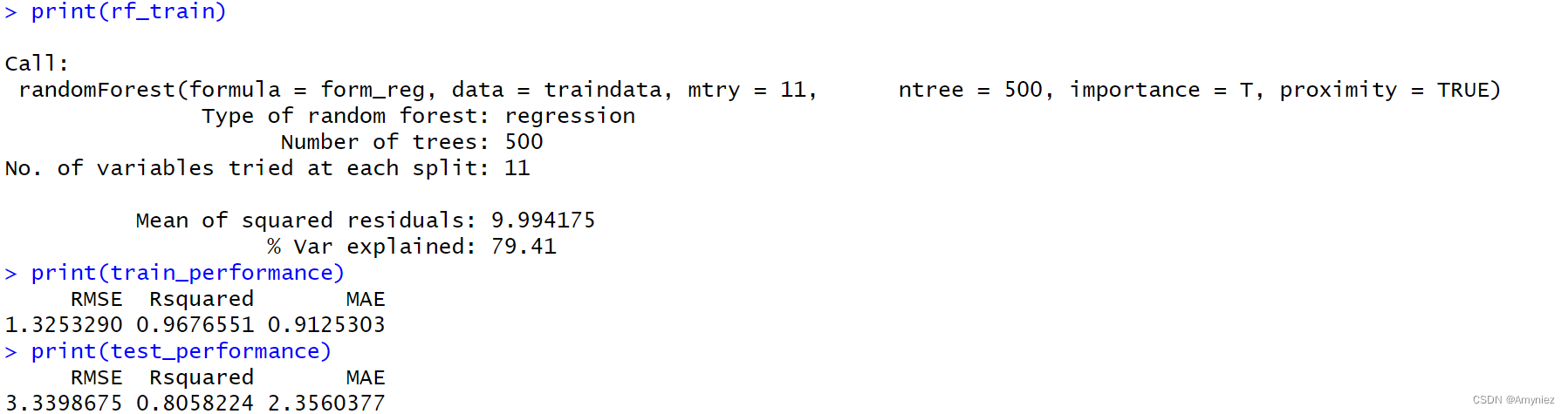
rf_train<-randomForest(form_reg,data=traindata,mtry=11,ntree=500,importance = T,proximity=TRUE)
plot(rf_train,main = "ERROR & TREES") #绘制模型误差与决策树数量关系图
运行结果:
6. 查看变量重要性并绘图展示
#### 变量重要性
importance<-importance(rf_train) ##### 绘图法1
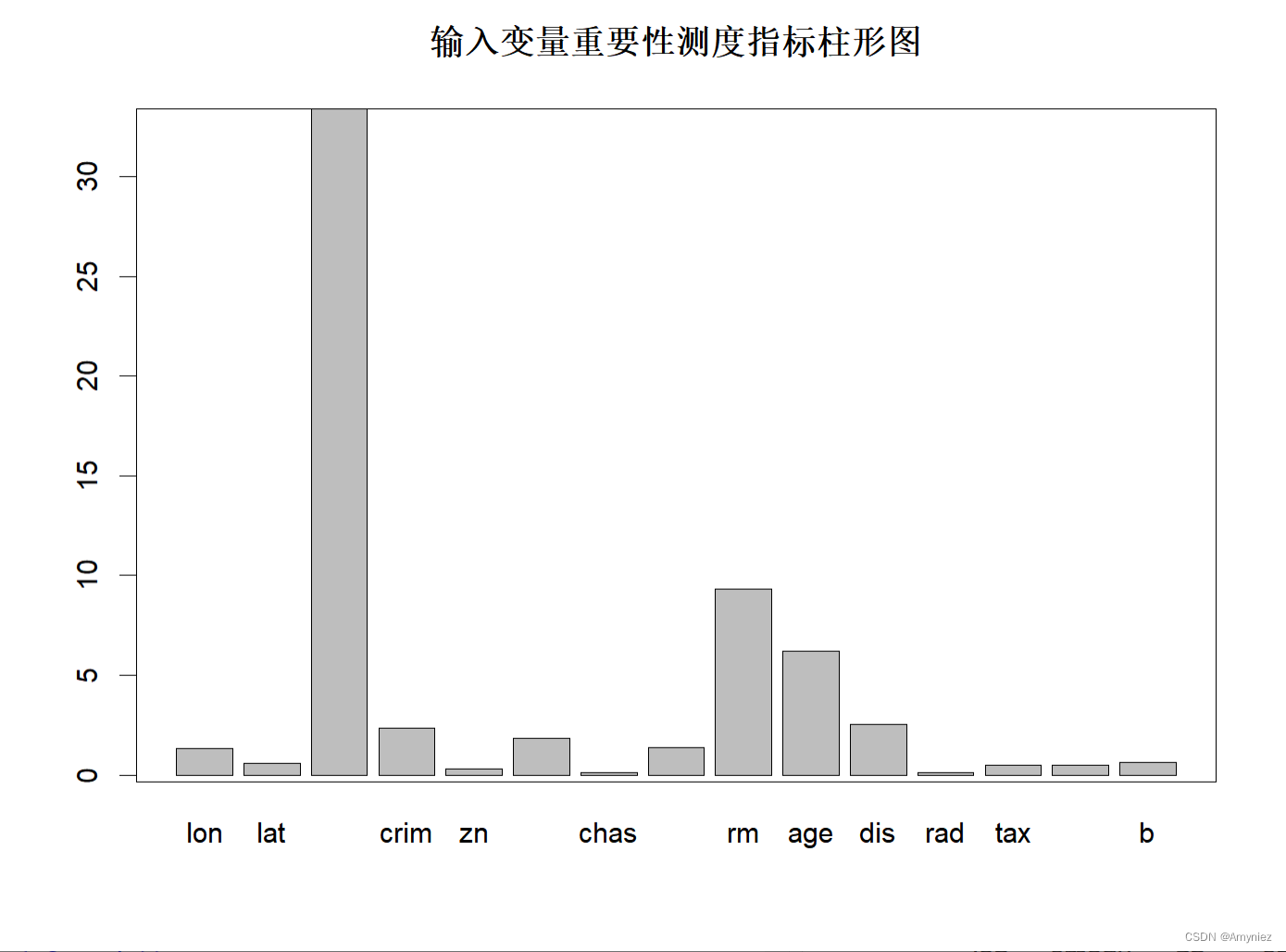
barplot(rf_train$importance[,1],main="输入变量重要性测度指标柱形图")
box()
重要性展示:
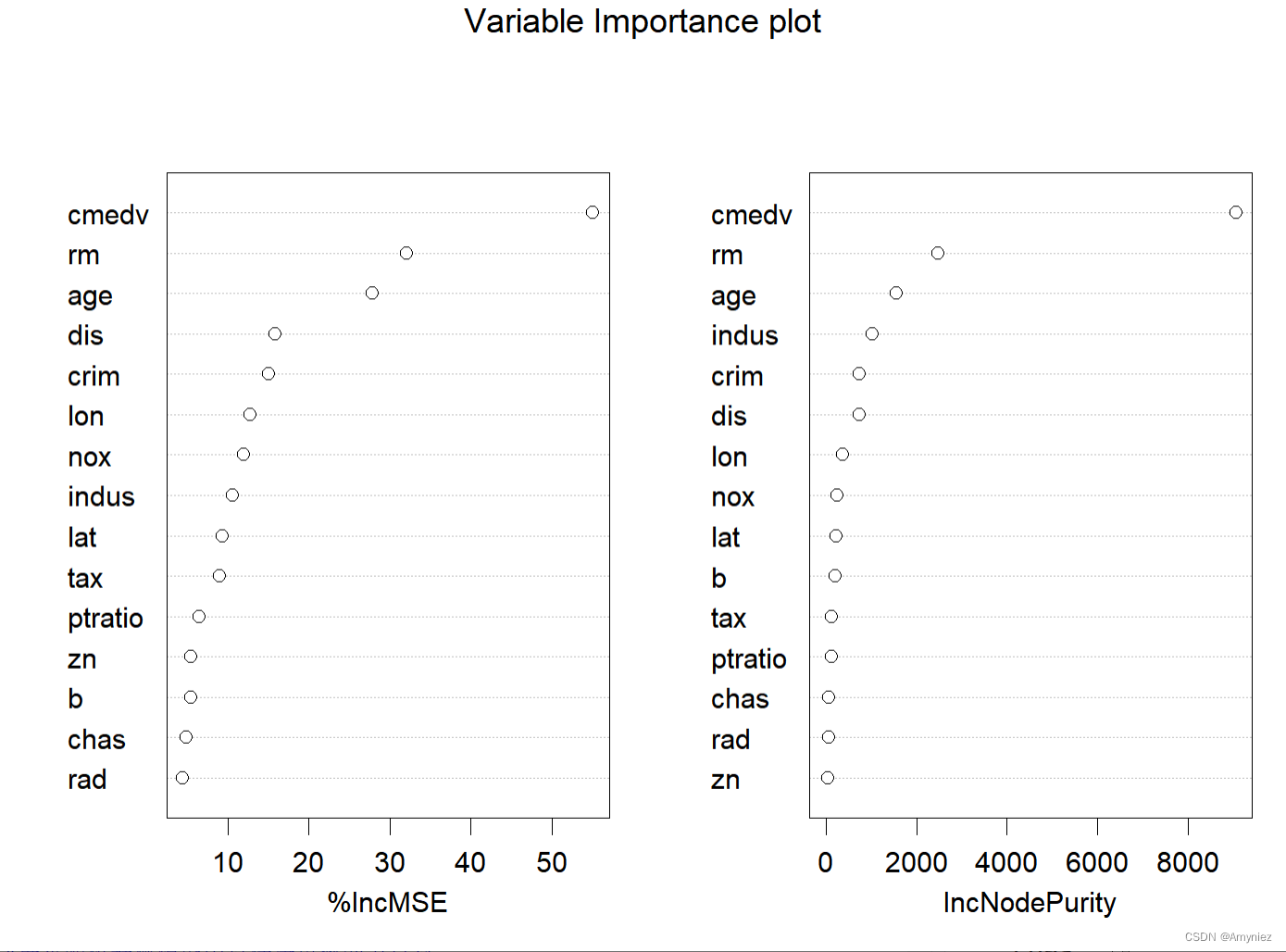
##### 绘图法2
varImpPlot(rf_train,main = "Variable Importance plot")
varImpPlot(rf_train,main = "Variable Importance plot",type = 1)
varImpPlot(rf_train,sort=TRUE,n.var=nrow(rf_train$importance),main = "Variable Importance plot",type = 2) # 基尼系数
hist(treesize(rf_train)) #展示随机森林模型中每棵决策树的节点数
max(treesize(rf_train));
min(treesize(rf_train))
“%IncMSE” 即increase in mean squared error,通过对每一个预测变量随机赋值,如果该预测变量更为重要,那么其值被随机替换后模型预测的误差会增大。“IncNodePurity”即increase in node purity,通过残差平方和来度量,代表了每个变量对分类树每个节点上观测值的异质性的影响,从而比较变量的重要性。两个指示值均是判断预测变量重要性的指标,均是值越大表示该变量的重要性越大,但分别基于两者的重要性排名存在一定的差异。
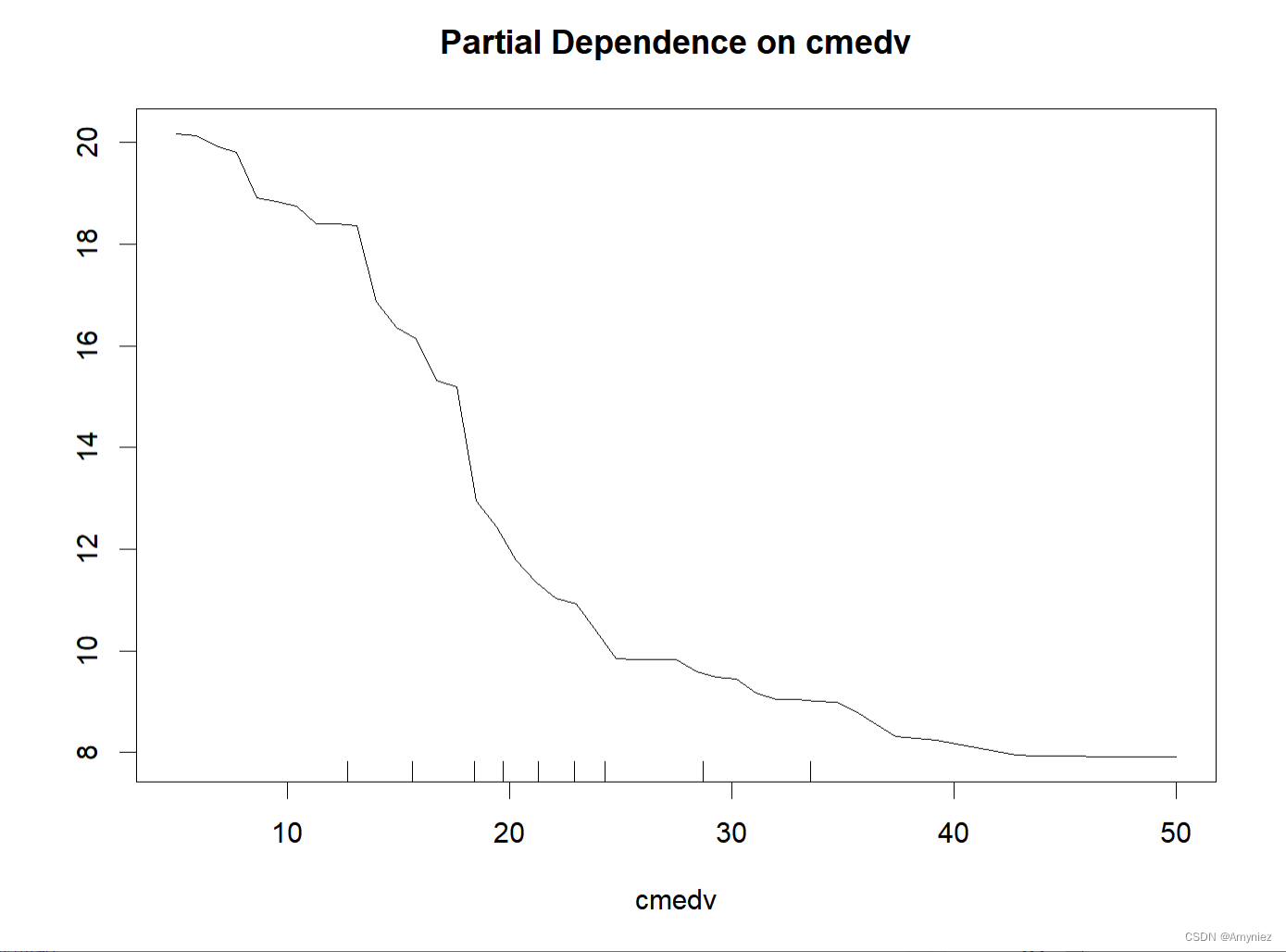
7. 偏依赖图:Partial Dependence Plot(PDP图)
部分依赖图可以显示目标和特征之间的关系是线性的、单调的还是更复杂的
缺点: 部分依赖函数中现实的最大特征数是两个,这不是PDP的错,而是2维表示(纸或屏幕)的错,是我们无法想象超过3维的错。
partialPlot(x = rf_train,pred.data = traindata,x.var = cmedv
)
PDP图:
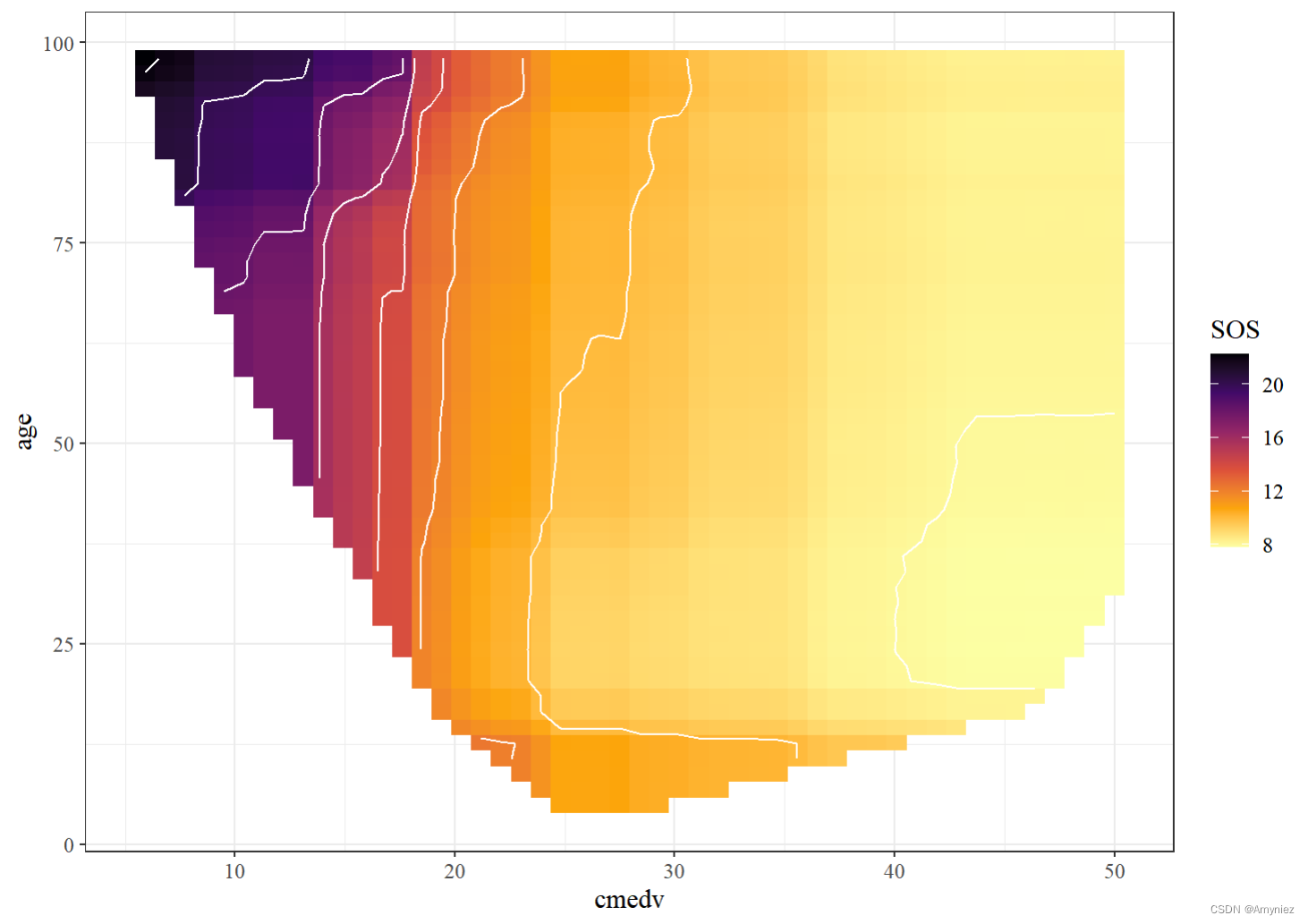
rf_train %>%partial(pred.var = c("cmedv", "age"), chull = TRUE, progress = TRUE) %>%autoplot(contour = TRUE, legend.title = "SOS",option = "B", direction = -1) + theme_bw()+theme(text=element_text(size=12, family="serif"))
交互结果展示:
#预测与指标的关系散点图
plot(lstat ~ cmedv, data = traindata)
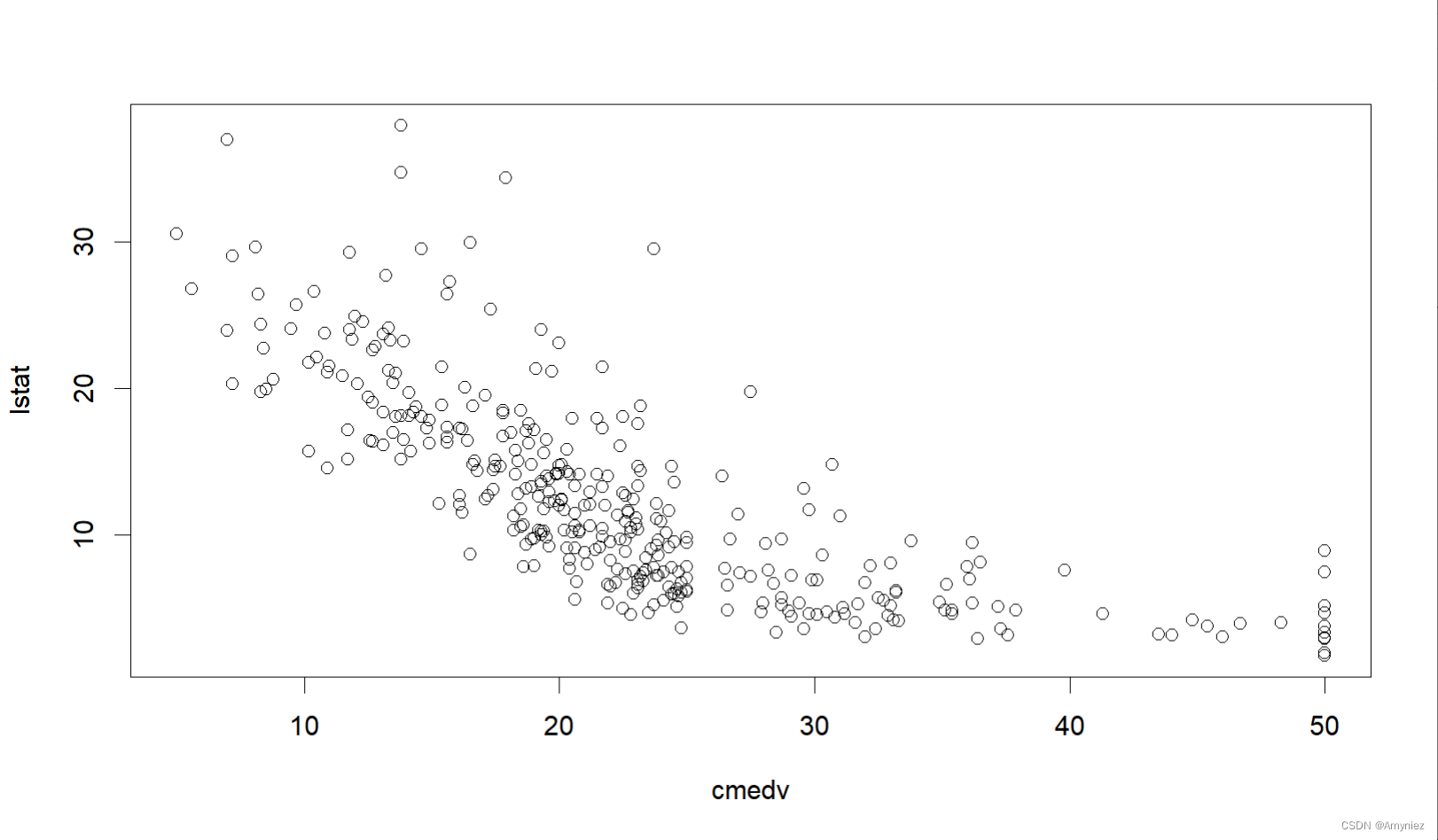
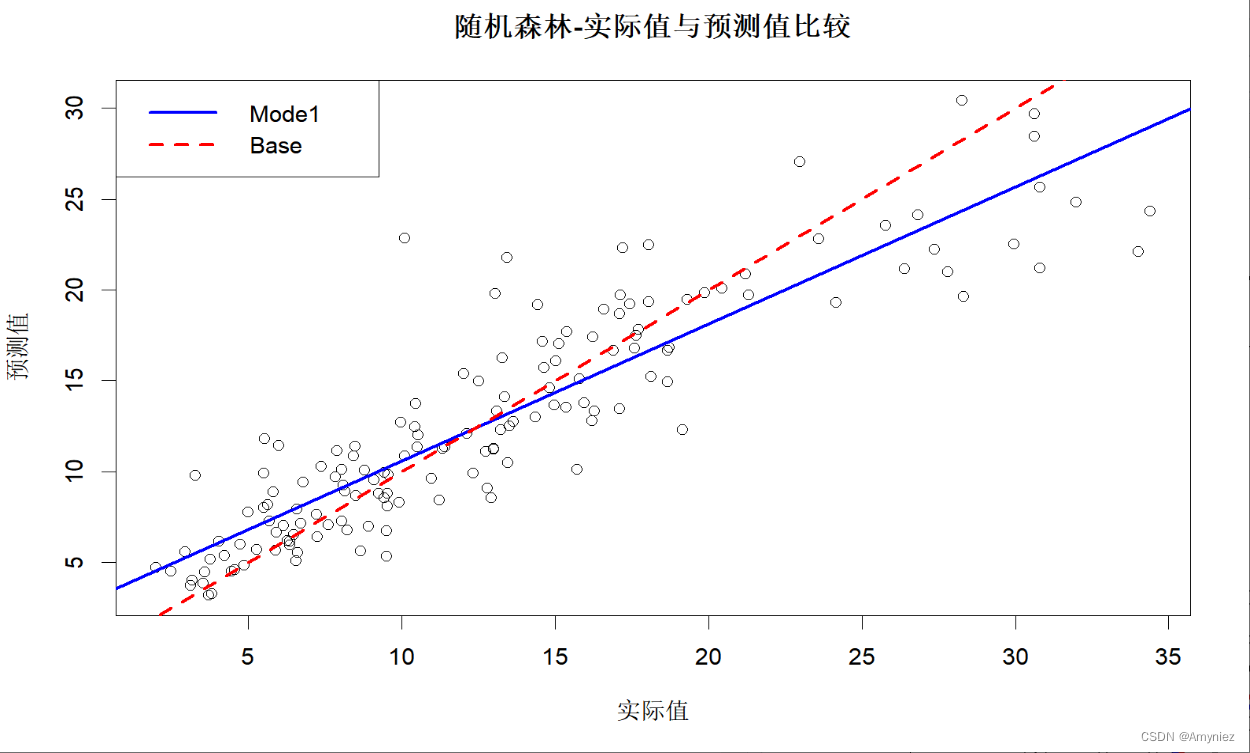
8. 训练集预测结果
#图示训练集预测结果
plot(x = traindata$lstat,y = trainpred,xlab = "实际值",ylab = "预测值",main = "随机森林-实际值与预测值比较"
)trainlinmod <- lm(trainpred ~ traindata$lstat) #拟合回归模型
abline(trainlinmod, col = "blue",lwd =2.5, lty = "solid")
abline(a = 0,b = 1, col = "red",lwd =2.5, lty = "dashed")
legend("topleft",legend = c("Mode1","Base"),col = c("blue","red"),lwd = 2.5,lty = c("solid","dashed"))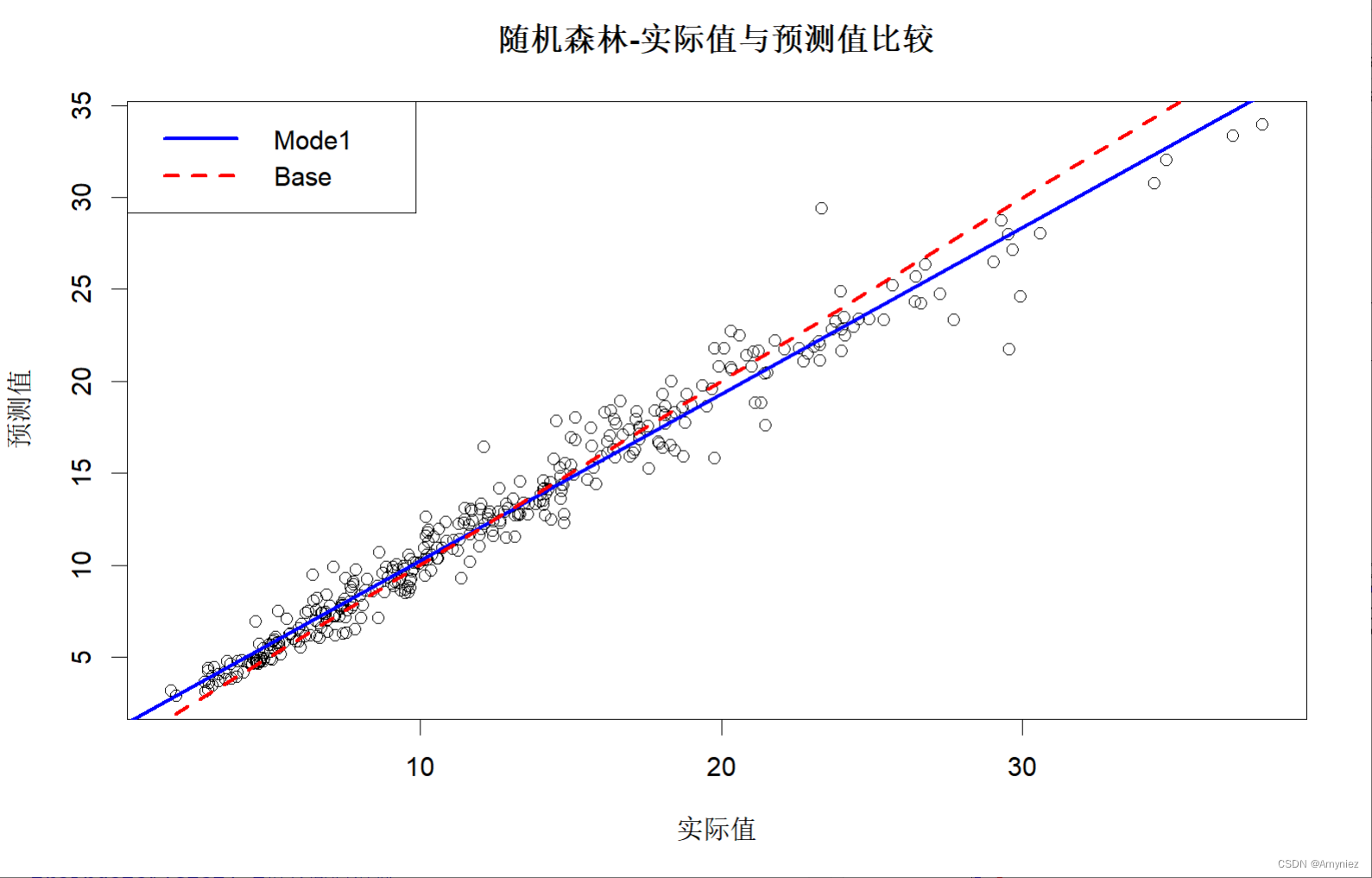
#测试集预测结果
testpred <- predict(rf_train,newdata = testdata)
#测试集预测误差结果
defaultSummary(data.frame(obs = testdata$lstat,pred = testpred))
#图示测试集结果
plot(x = testdata$lstat,y = testpred,xlab = "实际值",ylab = "预测值",main = "随机森林-实际值与预测值比较"
)
testlinmod <- lm(testpred ~ testdata$lstat)
abline(testlinmod, col = "blue",lwd =2.5, lty = "solid")
abline(a = 0,b = 1, col = "red",lwd =2.5, lty = "dashed")
legend("topleft",legend = c("Mode1","Base"),col = c("blue","red"),lwd = 2.5,lty = c("solid","dashed"))